
易培珊,博士,特聘研究员,博士生导师。入选9999js金沙老品牌双百人才工程计划(2020年),四川省青年人才(2021年),国家高层次人才计划青年人才(2024年)。现任Plant Cell Reports期刊编委,中国细胞生物会细胞结构与细胞行为分会委员,四川省植物学会理事,四川省细胞生物学会理事。长期从事细胞极性与细胞行为机制研究。以第一作者(含共同)和通讯作者身份在Nature Communications,Current Biology,Plant Cell,New Phytologist,Plant Biotechnology Journal等期刊发表SCI论文十余篇。研究受到人类前沿科学研究项目(Human Frontier Science Program),国家自然科学基金, 四川省青年人才项目等支持。主讲本科生《细胞生物学(双语)》和《细胞生物学实验》,研究生《植物细胞生物学》三门课程。
联系方式:yipeishan@scu.edu.cn
联系地址:四川省成都市武侯区一环路南一段24号9999js金沙老品牌A501,610064
实验室主页:https://psyi.github.io/【网站Contact目录下有硕士/博士名额剩余情况,随时更新】
个人主页:https://orcid.org/0000-0003-4710-8089
学习和工作经历:
2007.09 – 2011.07 北京大学,本科
2011.09 – 2014.07 中国科学院大学,中科院生物物理研究所,硕士,导师:欧光朔
2014.09 – 2017.06 清华大学,博士,导师:欧光朔
2017.10 – 2020.09 名古屋大学(日本),博士后,导师:五島 剛太(Gohta Goshima)
2020.12 – 至今 9999js金沙老品牌,9999js金沙老品牌,特聘研究员
研究方向:
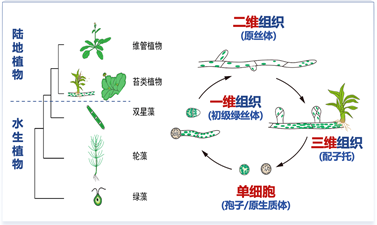
课题组长期从事细胞生长、分裂与分化调控机制研究。近年以小立碗藓(Physcomitrium patens)为对象,揭示小G蛋白信号在细胞极性生长和不对称分裂中的重要作用,提出植物细胞不对称分裂的通用模型;在苔藓中建立基于CRISPR/Cas9的突变敲入技术。小立碗藓是早期陆生植物的代表性分支之一,生长方式包括二维线性(原丝体)和三维多轴向(配子托)形式。课题组目前关注苔藓植物形态发生和体轴建立的细胞分子机制和基本原理,探讨植物从水生向陆生过度阶段,细胞结构、行为与发育模式的适应性改变。研究手段包括基因敲除与敲入、活体细胞成像、体外生化分析等,研究方向包括:
1. 植物细胞极性与不对称分裂
2. 植物细胞的生长、形态发生和维持
3. 细胞骨架和胞内运输
4. 遗传工具的开发与应用
基金项目:
2018 – 2020 The Human Frontier Science Program Long-Term Fellowship
2022 – 2023 9999js金沙老品牌“从0到1”创新研究项目
2022 – 2024 国家自然科学基金青年基金
2023 – 2025 9999js金沙老品牌科技领军人才培育项目
2023 – 2026 国家自然科学基金面上项目
发表文章:(#共同一作,*通讯作者)
Tian, H., Lyu, R. and Yi, P.* (2024). Crosstalk between Rho of Plants GTPase signalling and plant hormones. J Exp Bot 75: 3778-3796.
Ruan, J., Yin, Z. and Yi, P.* (2024). Effects of fluorescent tags and activity status on the membrane localization of ROP GTPases. Plant Signal Behav 19: 2306790.
Ruan, J.#, Lai, L.#, Ou, H., and Yi, P.* (2023). Two subtypes of GTPase-activating proteins coordinate tip growth and cell size regulation in Physcomitrium patens. Nat Commun 14:7084.
Lai, L., Ruan, J., Xiao, C., and Yi, P.* (2023). The putative myristoylome of Physcomitrium patens reveals conserved features of myristoylation in basal land plants. Plant Cell Rep 42:1107-1124.
Ou, H., and Yi, P.* (2022). ROP GTPase-dependent polarity establishment during tip growth in plants. New Phytol 236:49-57.
Ruan, J. and Yi, P.* (2022). Exogenous 6-benzylaminopurine inhibits tip growth and cytokinesis via regulating actin dynamics in the moss Physcomitrium patens. Planta 256:1.
Yi, P.* and Goshima, G. (2022). Division site determination during asymmetric cell division in plants. Plant Cell 34:2120–2139.
Li, D., Liu, Y., Yi, P., Zhu, Z., Li, W., Zhang, Q.C., Li, J.B., and Ou, G. (2021). RNA editing restricts hyperactive ciliary kinases. Science 373:984-991.
Yi, P.* and Goshima, G. (2020). Rho of Plants GTPases and cytoskeletal elements control nuclear positioning and asymmetric cell division during Physcomitrella patens branching. Curr Biol 30:2860-2868.
Yi, P.* and Goshima, G. (2020). Transient cotransformation of CRISPR/Cas9 and oligonucleotide templates enables efficient editing of target loci in Physcomitrella patens. Plant Biotechnol J 18:599-601.
Yi, P.* and Goshima, G.* (2018). Microtubule nucleation and organization without centrosomes. Curr Opin Plant Biol 46:1-7.
Yi, P., Xie, C., and Ou, G.* (2018). The kinases male germ cell-associated kinase and cell cycle-related kinase regulate kinesin-2 motility in Caenorhabditis elegans neuronal cilia. Traffic 19:522-535.
Yi, P., Li, W.J., Dong, M.Q., and Ou, G.* (2017). Dynein-Driven Retrograde Intraflagellar Transport Is Triphasic in C. elegans Sensory Cilia. Curr Biol 27:1448-1461.
Li, W.#, Yi, P.#, and Ou, G.* (2015). Somatic CRISPR-Cas9-induced mutations reveal roles of embryonically essential dynein chains in Caenorhabditis elegans cilia. J Cell Biol 208:683-692.
Yi, P., Li, W., and Ou, G.* (2014). The application of transcription activator-like effector nucleases for genome editing in C. elegans. Methods 68:389-396.
Cheng, Z.#, Yi, P.#, Wang, X., Chai, Y., Feng, G., Yang, Y., Liang, X., Zhu, Z., Li, W., and Ou, G.* (2013). Conditional targeted genome editing using somatically expressed TALENs in C. elegans. Nat Biotechnol 31:934-937.
Feng, G.#, Yi, P.#, Yang, Y.#, Chai, Y.#, Tian, D.#, Zhu, Z., Liu, J., Zhou, F., Cheng, Z., Wang, X., et al. (2013). Developmental stage-dependent transcriptional regulatory pathways control neuroblast lineage progression. Development 140:3838-3847.




